Example Virtual Training Projects (VTPs)
VTPs Utilize:
- Bioinformatics, computational biology and/or data science methods. Examples include: Data QC, Filtering, Normalization, Regression, Clustering, Biomarker Identification, Dimensionality Reduction, and Data Visualization (e.g. violin plots, scatterplots, heatmapts), Gene Set Enrichment/Pathway Analysis, Bulk RNAseq, Single-cell Transcriptomics/Epigenomics, Social Mobility Analysis, and Machine Learning.
- Open-source programming languages (e.g. Linux, R, Python) so participants can use acquired skills after finishing a VTP without purchasing cost-prohibitive software.
- Publicly available datasets so participants can continue to utilize the data if desired.
- Alignment with established analysis guidelines. In each VTP, we examine how the performed tasks align with, deviate from, or augment the standard practices for a given analysis type. When no consensus exists, we explore and discuss the varying perspectives.
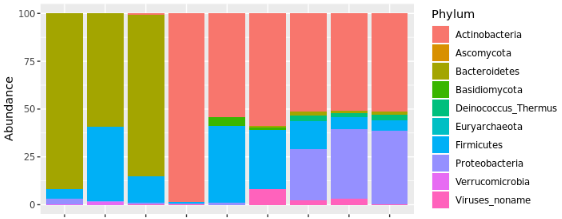
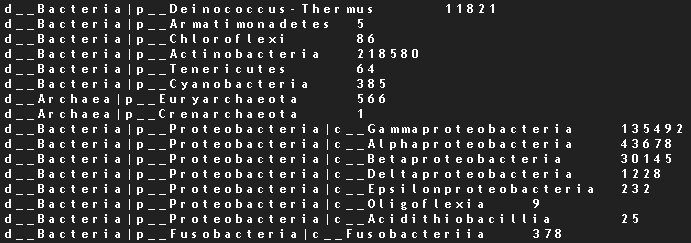
Metagenomics + Microbial Surveillance
Collaboration with Dr. David Danko, Mason Lab, Cornell Medical Center
Goal
Characterize mass-transit environmental microbiome samples using metagenomic analysis.
Domains
Linux, Data Analysis & Visualization in R, Metagenomics, Principal Component Analysis, Kraken (bioinformatics tool for metagenomic characterization/quantification)
Data Source
Danko, D., Bezdan, D. et al. Global Genetic Cartography of Urban Metagenomes and Anti-Microbial Resistance. Preprint on bioRxiv.
Danko, D., Bezdan, D. et al. A global metagenomic map of urban microbiomes and antimicrobial resistance. Cell. New York Times coverage.
Detailed Descriptions:
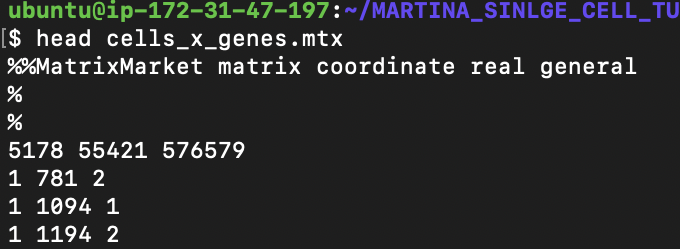
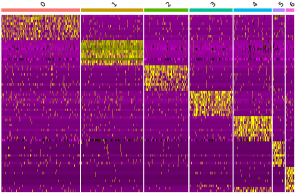
Single-cell Transcriptomics + Lung Cell Characterization
Collaboration with Dr. Martina Bradic, Memorial Sloan Kettering Cancer Center
Goal
Transcriptomically profile the aging mouse lung using single-cell RNA-sequencing (scRNA-seq) analysis
Domains
Seurat, Linux, R, Library QC, Data filtering/clustering/PCA/t-SNE, Cell-type Assignment, Gene Expression, Drop-seq, 10x (optional additional dataset)
Data Sources
Angelidis, I., Simon, L.M., Fernandez, I.E. et al. An atlas of the aging lung mapped by single cell transcriptomics and deep tissue proteomics. Nature Communications 2019. Preprint on bioRxiv.
Optional additional 10x dataset:
O’Koren, E., Yu, C. et al. Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration. Immunity.
Detailed Descriptions:
Goal
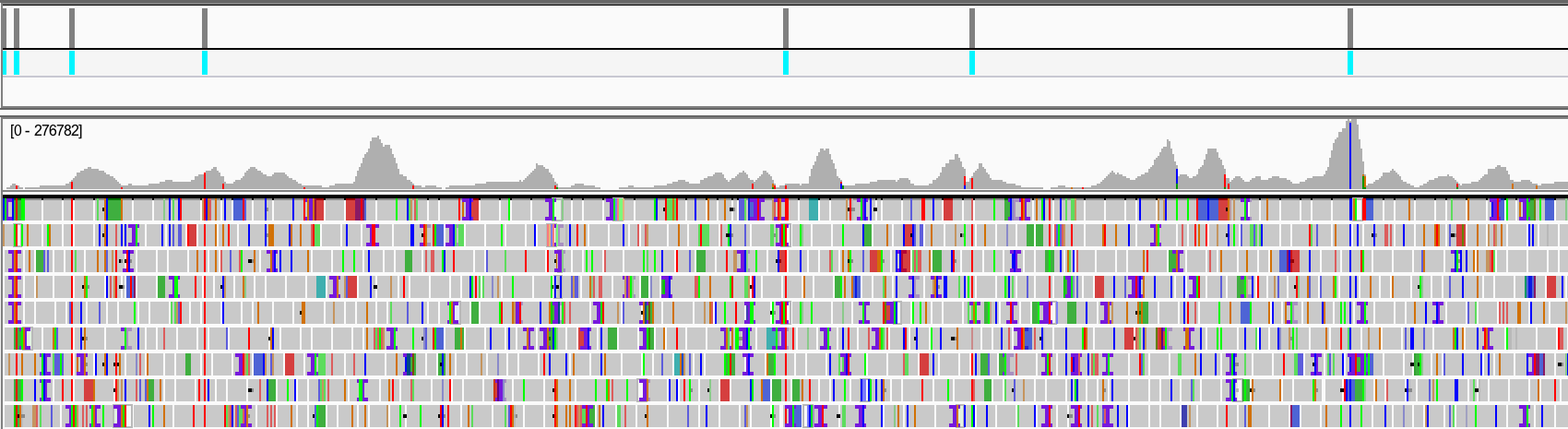
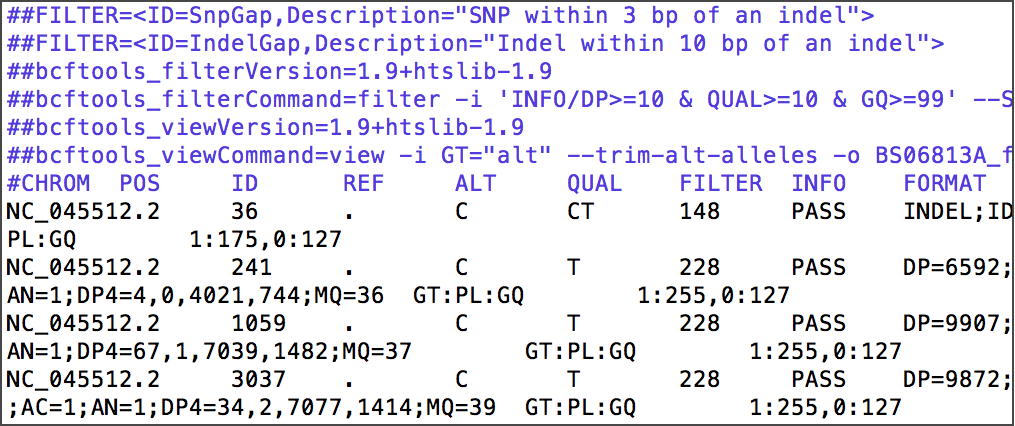
Characterize nucleotide and amino acid variants from SARS-CoV-2+ patient samples
Domains
Linux, Bowtie2, SAMtools, BCFtools, IGV Genome Browser, GISAID
Data Source
Maurano et al. Sequencing identifies multiple early introductions of SARS-CoV-2 to the New York City Region. Genome Research 2020. Preprint on MedRxiv. New York Times coverage.
Detailed Descriptions:
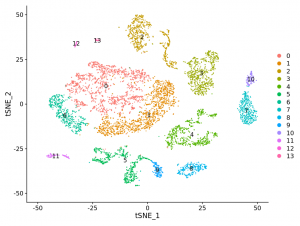
Single-cell Transcriptomics + Neuron Characterization in Habenula
Collaboration with Asst. Prof. Mike Wallace, Boston University School of Medicine (Formerly, Harvard Medical School)
Goal
Transcriptomically profile the murine habenula using single-cell RNA-sequencing (scRNA-seq) analysis.
Domains
Cellular/molecular neuroscience, Seurat, Linux, R, Library QC, Data filtering/clustering/PCA/t-SNE, Cell-type Assignment, Gene Expression, 10x (optional additional dataset)
Data Sources
Wallace et al. Anatomical and single-cell transcriptional profiling of the murine habenular complex. eLife 2020.
Optional additional 10x dataset:
O’Koren, E., Yu, C. et al. Microglial Function Is Distinct in Different Anatomical Locations during Retinal Homeostasis and Degeneration. Immunity.
Detailed Descriptions:

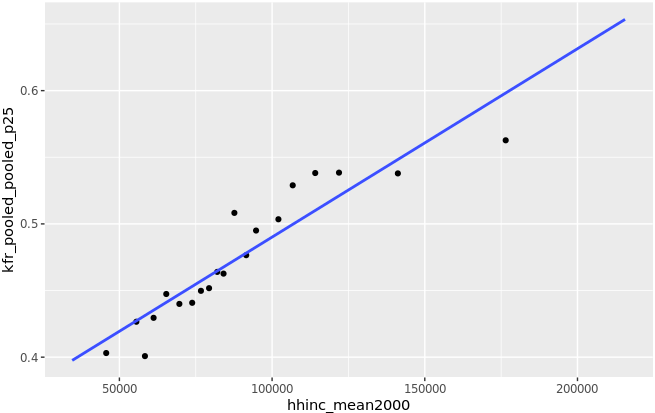
Goal
Analyze and explore the childhood roots of social mobility at the community level.
Domains
Data Analysis in R, Data Visualization, Regression & Correlation Analysis
Data Source
Chetty et al. The Opportunity Atlas: Mapping the Childhood Roots of Social Mobility. NBER. Manuscript on Opportunity Insights. New York Times coverage.
Detailed Descriptions:
Single-cell Epigenomics + Neurodegenerative Disorder Elucidation
Goal
Profile the epigenomic state of human brain cells using single-cell Assay for Transposase-Accessible Chromatin sequencing (scATAC-seq) analysis and explore their implications for Alzheimer’s and Parkinson’s diseases.
Domains
Cellular/Molecular Neuroscience, Neurodegenerative Disorders, Epigenetic State Analysis, Single-cell Epigenomics, Linux, R
Data Source
M. Ryan Corces et al. Single-cell epigenomic analyses implicate candidate causal variants at inherited risk loci for Alzheimer’s and Parkinson’s diseases. Nature Genetics.
Detailed Descriptions:
Coming Soon
Stem Cell Biology + Pathway Analysis + Oncology
Goal
Conduct single-cell transcriptomics and pathway analysis to elucidate mechanisms underlying colorectal cancer and stem-cell Biology/regeneration.
Domains
Cellular/Molecular Biology, Cancer Genetics, Single-cell Trancriptomics, Pathway Analysis, Linux, R
Data Source
Priscilla Cheung, Jordi Xiol et al. Regenerative Reprogramming of the Intestinal Stem Cell State via Hippo Signaling Suppresses Metastatic Colorectal Cancer. Cell Stem Cell.
Detailed Descriptions:
- Research Staff · Postdoc · Graduate Student Track
- Undergraduate · High School Student Track
Epigenetic Analysis + Tumorigenesis Initiation
Goal
Epigenetically profile functionally perturbed mouse model pancreatic tissue to explore how gene mutations and tissue damage combine to promote a chromatin structure that initiates tumor emergence.
Domain
Cellular/Molecular Biology, Cancer Genetics, Epigenetic State Analysis, Single-cell Epigenomics, Linux, Python
Data Source
Alonso-Curbelo, D., Ho, YJ., Burdziak, C. et al. A gene–environment-induced epigenetic program initiates tumorigenesis. Nature.
Detailed Descriptions:
- Research Staff · Postdoc · Graduate Student Track
- Undergraduate · High School Student Track
Cardiogenesis + Cardiovascular Defects + Cell Trajectory Analysis
Goal
Analyze the developmental trajectories of cardiac cells from healthy and disease-model mice to elucidate the molecular mechanisms of cardiovascular defects.
Domains
Cellular/Molecular Biology, Developmental Biology, Cardiovascular Biology, Pseudotime analysis. Linux, R
Data Source
de Soysa, T.Y., Ranade, S.S., Okawa, S. et al. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature.
Detailed Descriptions:
- Research Staff · Postdoc · Graduate Student Track
- Undergraduate · High School Student Track
Spatial Transcriptomics + Amyotrophic Lateral Sclerosis
Goal
Using spatial transcriptomics, analyze control and amyotrophic lateral sclerosis (ALS) model mice at presymptomatic, onset, symptomatic, and end-stage time points to elucidate the underlying molecular mechanisms in ALS.
Domains
Cellular/Molecular Neuroscience, Neurodegenerative Disorders, Spatial Transcriptomics Analysis, Linux, Python
Data Source
Silas Maniatis, Tarmo Äijö, Sanja Vickovic et al. Spatiotemporal dynamics of molecular pathology in amyotrophic lateral sclerosis. Science.
Detailed Descriptions:
- Research Staff · Postdoc · Graduate Student Track
- Undergraduate · High School Student Track
Single-cell Transcriptomics + Stem Cell Microenvironment
Goal
Characterize the transcriptional landscape of bone-marrow microenvironment at single-cell resolution in homeostasis and under stress-induced haematopoiesis.
Domains
Cellular/Molecular Biology, Stem-cell Microenvironment, Hematology, scRNAseq, Bulk RNA-seq (optional), Linux, R
Data Source
Tikhonova, A.N., Dolgalev, I., Hu, H. et al. The bone marrow microenvironment at single-cell resolution. Nature.
Detailed Descriptions:
- Research Staff · Postdoc · Graduate Student Track
- Undergraduate · High School Student Track
DNA Methylation State + Embryonic Stem Cell Biology + CRISPR Screens
Goal
Using data from a genome-wide CRISPR-Cas9 screen in human embryonic stem cells, characterize and validate top-hit DNA methylation regulators.
Domains
Cellular/Molecular Biology, CRISPR Screens, Methylation Analysis, Pathway Analysis, Linux, R
Data Source
Gary Dixon, Heng Pan, et al. QSER1 protects DNA methylation valleys from de novo methylation. Science.
Detailed Descriptions:
- Research Staff · Postdoc · Graduate Student Track
- Undergraduate · High School Student Track
Spatial Microbiome Analysis + Tumor Progression
Goal
Investigate the organized spatial distribution of intratumoral microbiota to interrogate its impact on tumor progression.
Domains
16S-sequencing, spatial transcriptomics, GATK/PathSeq, Linux, Python.
Data Source
Galeano Niño, J.L., Wu, H., LaCourse, K.D. et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature.
Detailed Descriptions:
- Research Staff · Postdoc · Graduate Student Track
- Undergraduate · High School Student Track
Please contact us at [email protected] for more information about our VTPs.